Background: In this investigation, we explore the literature regarding neuroregeneration from the 1700s to the present. The regeneration of central nervous system neurons or the regeneration of axons from cell bodies and their reconnection with other neurons remains a major hurdle. Injuries relating to war and accidents attracted medical professionals throughout early history to regenerate and reconnect nerves. Early literature till 1990 lacked specific molecular details and is likely provide some clues to conditions that promoted neuron and/or axon regeneration. This is an avenue for the application of natural language processing (NLP) to gain actionable intelligence. Post 1990 period saw an explosion of all molecular details. With the advent of genomic, transcriptomics, proteomics, and other omics—there is an emergence of big data sets and is another rich area for application of NLP. How the neuron and/or axon regeneration related keywords have changed over the years is a first step towards this endeavor.
Methods: Specifically, this article curates over 600 published works in the field of neuroregeneration. We then apply a dynamic topic modeling algorithm based on the Latent Dirichlet allocation (LDA) algorithm to assess how topics cluster based on topics.
Results: Based on how documents are assigned to topics, we then build a recommendation engine to assist researchers to access domain-specific literature based on how their search text matches to recommended document topics. The interface further includes interactive topic visualizations for researchers to understand how topics grow closer and further apart, and how intra-topic composition changes over time.
Conclusions: We present a recommendation engine and interactive interface that enables dynamic topic modeling for neuronal regeneration.
The scientific literature on neuro-regeneration and reinnervation began in the 18th century (1). Since then, hundreds of articles have been published on topics ranging from key regeneration genes (2-4) to the dynamics of remyelination (5). This knowledge corpus is vast and dynamic as new insights are continually emerging from research across multiple continents and topics. Given this vast accumulating body of research, it is hard to know how this literature is evolving. For example, which are the emerging research topics? Which ones have died out? Answers to such questions are important to scholars who want to pursue further research in this area or physicians who want to update their knowledge.
The purpose of this paper is to describe the design and implementation of a computational system that analyses digitally curated past research and provides researchers with an interface to query the corpus and get recommended papers. Furthermore, the web-based interactive interface provides a visualization tool to examine the evolution of topics over time. Using our proposed system, researchers can find answers to narrow queries such as the molecules that play a role in nerve regeneration or broad queries such as the gaps in the literature.
Our computational system relies on the advances made in natural language processing (NLP). Specifically, we use dynamic topic modelling (DTM) (6) to extracts topics and track changes of topics over time. It also shows users how keywords and topics evolve. Beyond applying DTM, we created a novel user interface for researchers to apply the trained model to their data. Overall, we make the following contributions to the interface of neural regeneration and computation:
The remainder of this paper is organized as follows: we review the literature and discuss related contributions from prior authors. Then we specify our algorithm design and computational tools. Next, we detail our experiments and report our results. We discuss the impact of our methods and opportunities for future investigation and present conclusions.
Researchers have developed computational tools for other domains, such as lipidomics and metabolomics. These tools help them garner insights from large scale non-structured text data. For example, Schomburg first composed the BRENDA (7) database in 2002, aiming to create a relational database between enzymes, proteins, and their respective biochemical pathways. By integrating information from KEGG metabolic pathways to diseases and other biomedical concepts, the researchers compose a translational database for scientists to examine changing terms over time. In 2017, Schomburg extended BRENDA by parsing 2.6 million papers in the PubMed corpus, extracting unstructured word tokens, and adding relevant labels (e.g., gene and enzyme names). This work represents a growing trend in biomedical sciences: applying NLPnatural language processing and Big Data techniques to derive structured insights. While BRENDA is powerful, it does not focus on the Neuroregeneration domain and does not focus on how terms and topics in the literature change over time. More importantly, BRENDA is applicable to detailed structural analysis rather than the analysis of broad topics and themes.
Besides relational databases and extraction of words, computational approaches like NLP have found use in biosciences. Chen (8) developed a Bio-DTM to explore how the traditional Chinese medical herb, ginseng, is discussed in the literature over time. While the corpus covers a wide array of topics, the literature lacks domain-focused DTM algorithms related to neuroscience, specifically Neuroregeneration.
Relatedly, van Altena (9) applied Latent Dirichlet Allocation (LDA) analysis methods to the PubMed and PMC corpora to model specific topics in those fields. After filtering for stop words and ranking their terms, the authors generated word clouds and other visuals to extract specific literature themes ranging from systems and security to disease prevention. The authors optimized their methods by tuning their number of topic hyperparameters to minimize the Akaike Information Criterion (AIC) (10) to choose the best model for their task. Other authors (11-13) use topic coherence (14,15) or other performance metrics (16) for optimizing the model architecture. After optimizing their performance metric, van Altena reported results on the broad biomedical corpus, yet the results lack domain-specificity.
LDA has applications beyond summarizing topics in a corpus: Wang (17) used Bio-LDA to discover key topics in PubMed, and then utilized entropy between keywords to generate a semantic ontology linking gene, disease, and other biological terms in an undirected graph. Beyond ontologies, Hu (18) derived LDA embeddings for unsupervised style suggestions for Etsy users. Although the domain is not related to medicine, their work shows how LDA can be used for inference and suggesting new content for users.
As we show, most of the current literature for Big Data NLP in Biomedicine handles broad corpora like the PubMed databases and lacks domain-specificity. Our focus on curating a corpus specific to Neuroregeneration and applying LDA-topic extraction methods provides nuanced insights in an important biomedical domain. Moreover, by applying DTM methods, we show how the literature evolves, allowing researchers to identify promising avenues for future research.
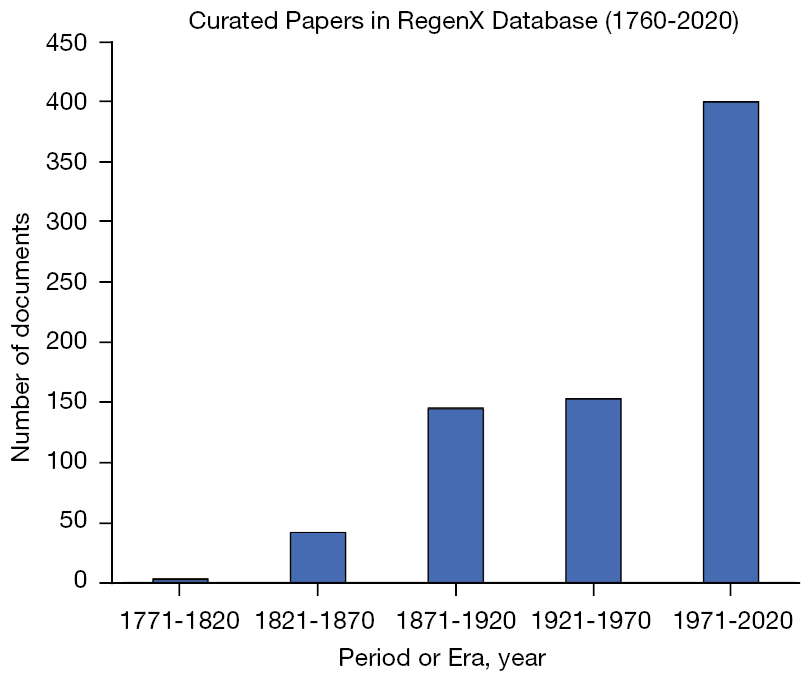
We began by curating the literature. To assemble this corpus, we searched in different databases, including Google Scholar and PubMed. We gathered 700 published articles and books within the Neuroregeneration domain and sorted them into bins based on their publication year. During our search, we also included papers in other languages, including French and German. In the 18th century, the scientific community was heterogeneous, and scientists often communicated their findings in their native languages. Despite this variation in language, the majority of the corpus is in English. Figure 1 illustrates the document distribution binned by time period. We chose to bin the entire corpus into 16 sequential time periods for training the DTM model.
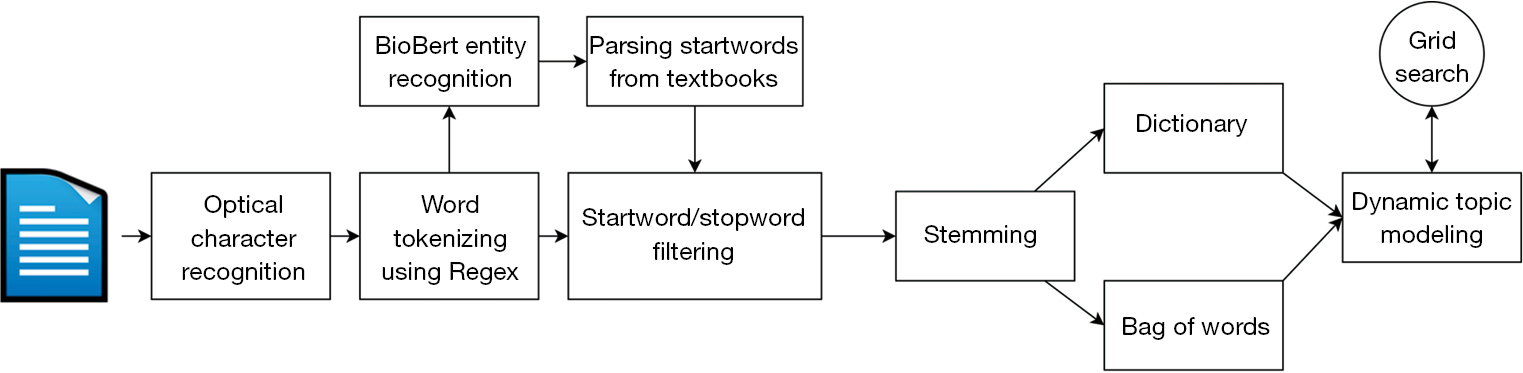
We used optical character recognition (OCR) technology to convert PDF format research papers into a machine-readable format that we can parse through. To achieve this end, we used the Adobe Document Cloud export PDF functionality (19), converting the .pdf files to .docx, which are parsed by specific Python libraries, as detailed below. When performing OCR, we discarded documents that the Document Cloud failed to process. These documents were primarily scanned images with complex figures or extremely large in disk space. Figure 2 details the overall process for handling the dataset.
After using OCR to convert the PDF documents into a usable string format, we preprocessed our text. We utilized simple regex to parse out words from non-letter characters. Additionally, we removed words lacking semantic meaning as stop words using the Python Natural Language Toolkit (NLTK)’s stop word list (20). Initial implementations yielded skewed results because the earlier periods contained some German, French, and other non-English articles. Thus, we also incorporated German, French, Dutch, and Spanish stop words to filter out words in languages beyond English.
To detect essential words in the literature, we assembled a collection of start words. We employed BioBERT (21) training tokens for this named entity recognition task. These tokens were derived from the BioCreative II Gene Mention corpus (22), the NCBI disease corpus (23), the CHEMDNER corpus of chemicals and drugs (24), and the Species-800 corpus (25), among others. We further included the indices of modern textbooks, including Netter’s Concise Neuroanatomy (26) and Bear’s Exploring the Brain (27), which contain significant named entities found throughout the corpus. Using these reference sources for start words created a clean filter when working with the data. Using our references for start words we removed these generic terms from the corpus, which allowed us to focus on domain-specific entities. Finally, we stemmed the words to remove redundancy, often found with word plurals and possessives. We chose to use Porter stemming (28) for this final preprocessing step.
We used the DTM algorithm, a time-series generative model for data collections over time. The algorithm builds off of Latent Dirichlet Allocation (LDA) which takes advantage of Bayesian probabilistic modelling of words in documents to define an underlying representation of topics in a document (29). The algorithm models each topic by an infinite mixture over an underlying set of topic probabilities to create a topic distribution for each research paper in our corpus. DTM extends this already powerful algorithm by separating the documents in our corpus by time slices, allowing us to model the evolution of topics over time. We analyzed the change in embeddings of each topic’s underlying probabilities over time by sequentially ordering our corpus. To extract the DTM topics, we relied on the Python Gensim package (30). We then used these topics to analyze our corpus in a variety of ways. DTM’s advantage lies in observing evolving entities and comparing documents from different periods with different word usage. In our experiments, we define the number of topics, T, and the number of time bins, B, as tunable parameters. We define these hyperparameters for selecting the optimal model. Further, we used CV Topic Coherence (31) and Domain expert validation as measures to compare our hyperparameters and evaluate the coherence of the topics produced. The systematic study of the configuration space of coherence measures is abbreviated as CV.
As noted above, there are many applications of LDA and DTM. We present a novel application of using our trained DTM to provide recommendations related to neural regeneration. Users can input the contents of their current research, abstracts of papers they are investigating, or anything that contains the topics for a researcher’s search query. We then preprocess the text following the same pipeline as the documents in our corpus. This is extremely important, as any differences in preprocessing will result in differences in interpretations from the model, yielding poor paper recommendations. The algorithm filters out words that are not start words, stems, tokenizes, and uses our dictionary to convert the tokens into a Bag of Words matrix. The DTM then calculates the log probability distribution of topics and compares this value to every research paper’s topic distribution in our corpus. We do this by calculating the Hellinger Distance between each of these topic distributions. In other words, we calculate how similar the topics in the text entered are to the topics in our set of research papers. We then return the research papers with the most similar topics. Below we discuss further the results of this implementation of DTM into a neural regeneration recommender system.
We ran a grid search to obtain the optimal hyperparameters for our DTM. Most notably, the tunable parameter with the most significant impact on this metric is the number of topics, T. Having too few topics will not give the recommendation algorithm sufficient flexibility to compare nuanced latent topics, both in the query entered by the user and the research papers in our corpus. For example, when the number of topics increases past (T>5) (T>5), we see a new topic emerge from our corpus that solely relates to the eye. Words such as ocular, retina, and vision comprise of this new topic. On the contrary, a T that is too high will result in incoherent topics, comprised of words with little semantic significance to Neuroregeneration researchers. Since the model’s primary purpose is to serve as part of a recommendation algorithm, we erred on the side of more topics, since the benefit of outputting more specialized topics outweighs the downside of producing incoherent topics. These incoherent topics would likely return low Hellinger Distances unless the user also entered incoherent text.
Most significantly, we used the CV Topic Coherence, which calculates the similarity of top words in a topic, to examine inter-topic similarity. Topic coherence measures the robustness of the topic distributions which allowed us to test different hyperparameters and measure their impact on our model. Figure 3 displays the grid search results below.
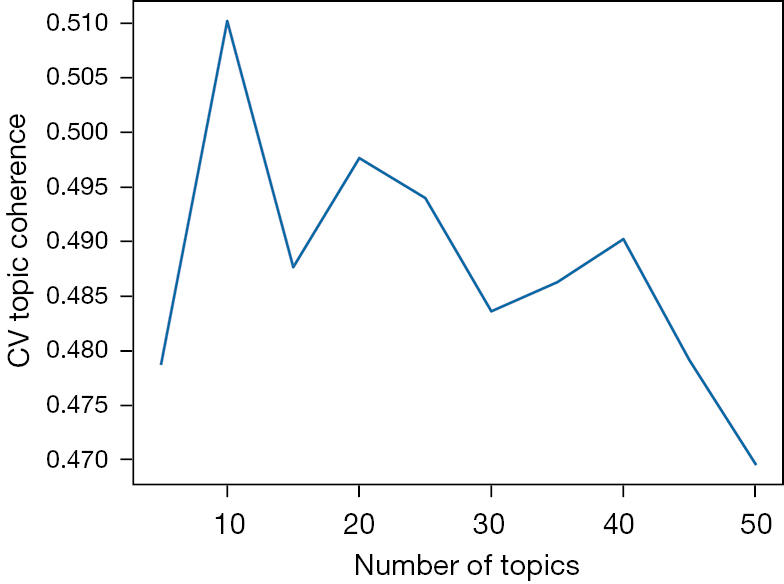
Ultimately, using grid search and topic coherence metrics alone were insufficient to decide the most optimal combination of hyperparameters. When deciding on overall topic quality, we also shared our discovered words and topics with Neuroscience undergraduate and graduate students and faculty at a large Southeastern University. We factored in their input to determine the optimal number of topics. To complete this qualitative assessment, we retrieved the top 20 terms for each topic over each binned time period. Through discussions with these domain experts, we chose to match the ten topics to the overall concepts found in Table 1. To assign topics, we surveyed four neurologists and neurology researchers independently for their summaries on the top 20 topic words. We present here the most salient responses.
| Topic number | Concept |
|---|---|
| 1 | Vision |
| 2 | Processes and activity |
| 3 | Anatomy |
| 4 | Cells |
| 5 | Growth and regeneration |
| 6 | Spinal cord |
| 7 | Disease |
| 8 | Movement |
| 9 | Optic nerve regeneration |
| 10 | CNS neuroanatomy |
CNS, central nervous system.
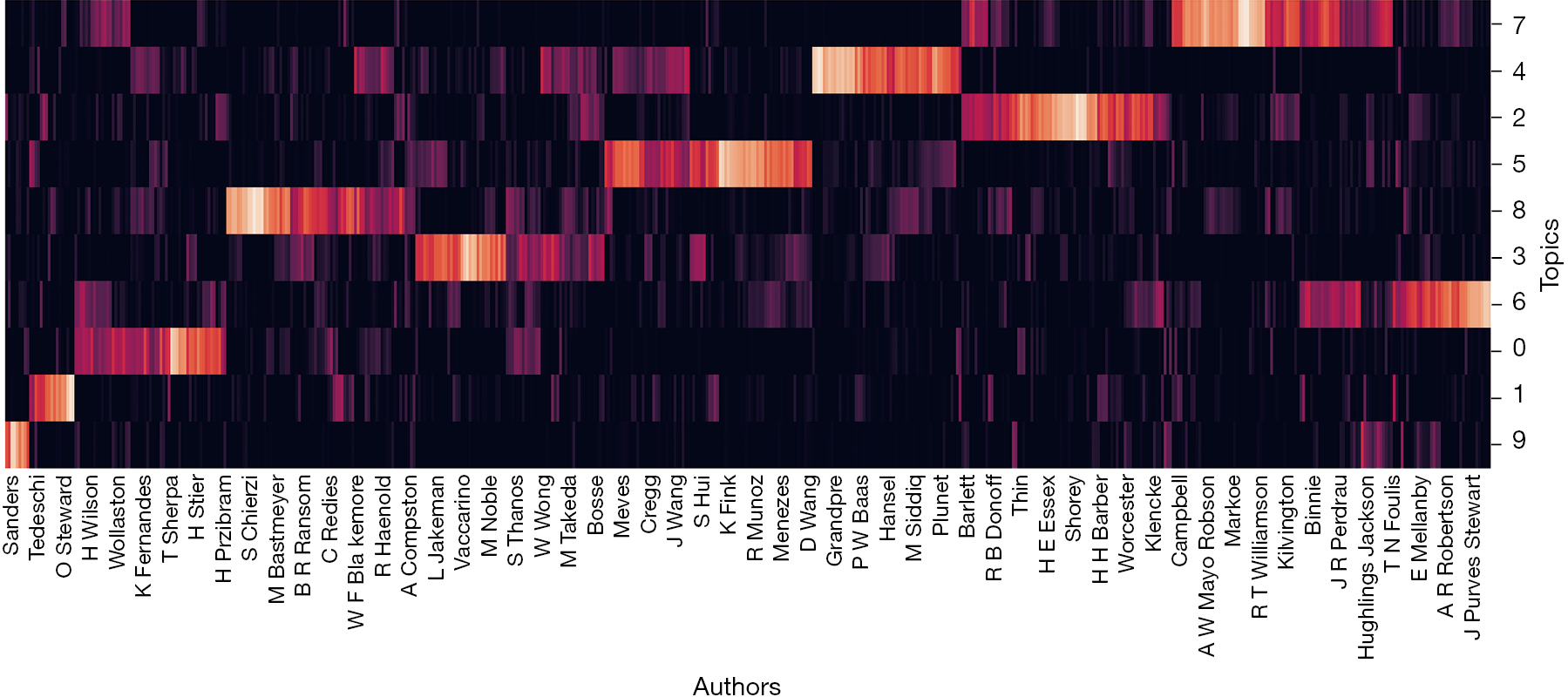
The DTM model also shows how authors cluster based on topic and shows salient patterns over the years. Figure 4 shows the results of authors found in the corpus and how they cluster together based on similar topics. We provide a key for the topics shown in the supplemental file (available online: https://cdn.amegroups.cn/static/public/10.21037aes-21-29-1.xls).
Another exciting application that arises from utilizing Bayesian probabilities to model Neuroregeneration research topics is the resulting log probabilities of keywords. In LDA and DTM, each word belongs to a topic, or even multiple topics. Topic assignment depends on probability of that word appearing in that topic. As a result of this, we can study how certain words rise and fall out of particular literature topics over time. In Figure 5, we see several keywords relating to neural regeneration research.
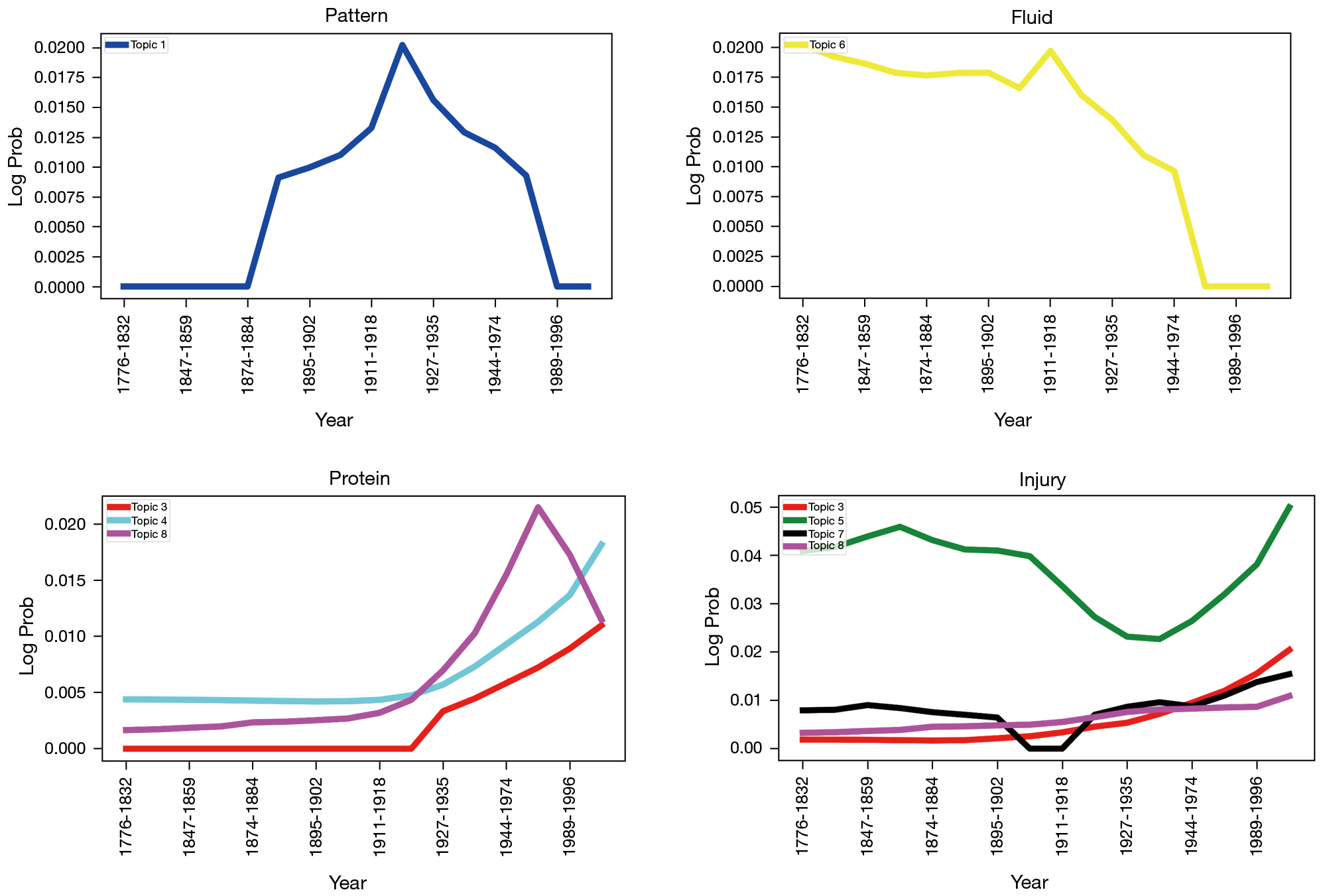
As mentioned above, our algorithm’s advantage lies in its robustness in recommending research papers from different periods that may have different words, syntax, and sentence structure. The model considers papers from different periods differing in everything but the latent topics embedded in that text for recommendation. Since neural regeneration is such a diverse subject, with a lengthy past, usage of this algorithm for recommendations results in finding research papers that would not ordinarily come up with similar searches on Google or PubMed. Additionally, by using Biobert entity and keyword filtering, we can fine-tune our DTM further to study the topics relating to our specific domain resulting in even stronger recommendations. Since we filtered the text in this way, we can create a flexible yet specific, set of topics closely aligned with the research domain.
The website also provides the user with the most important topic from our corpus related to their query, which presents another application. Perhaps the researcher is interested in gaining insights into what topics of regeneration their inputted text contains. In this manner, we include a powerful summarization feature on the website.
This article is the first to propose a computational approach to dynamic topic extraction from the corpus on Neuroregeneration literature. It serves as an entry point for future experiments using NLP to facilitate the broader key topic extraction from this research domain. We describe a text-mining pipeline for processing documents involving filtering techniques using BioBert. We also introduce a new approach for understanding topics, keywords, and subjects authors tend to write about over time. Finally, we implement all of the techniques above into a practical tool for the Neuroregeneration community to use involving our DTM that matches latent topics in the domain.
This paper’s limitations point to opportunities for further investigation. One such limitation is the link between the topics we derive from DTM. Specifically, to build an accurate knowledge system, we need to go beyond the bag-of-words approach and extract the text’s sequential information.
Finally, the DTM results herald further analysis and investigation. Our results are limited to analyzing temporal dynamics, yet geography is another crucial factor to ascertain when and where essential discoveries in the neuroscience field occurred. How do scientific discoveries’ time and location reflect geopolitical trends in history? These questions are open for future investigation, and forthcoming publications could focus on addressing these questions. We also hope to implement these future threads of research into additional practical functionality for researchers in the field to leverage.
This paper describes a computational system to investigate the dynamics of the Neuroregeneration literature. In particular, we curated a corpus of over 600 research papers and created a time-dependent dynamic topic model based on this corpus. This model is the backbone of a web application for researchers to visualize topics changing over time, to view how authors’ works cluster together, and to gain insights through an article recommendation system. By interpreting historical works, our applied tools advance neuroregeneration’s future development.