Myopia is an increasingly prevalent, complex trait influenced by numerous genetic and environmental factors. Whilst the increasing prevalence may be underlined by changing environmental pressures, the majority of trait variance is explained by genetic risk. In other words, the dispersion or variation in refractive error within individuals of a defined population is largely due to the effect of genes. This is supported by the crude but well-replicated dose-response association of one or more myopic parents and risk of myopia in their child (1-4), and to a better degree the high estimates of heritability for refractive error from twin studies (5-8). Theoretically, a genetic risk score containing all the genes contributing to myopia risk would explain the majority of the heritability and trait variance, whilst also providing a very useful predictive test. However, to date the identified genetic loci associated with refractive error explain only a small proportion of the estimated heritability, as discussed below.
Twin studies provide the unique opportunity to decompose a phenotype into the relative contribution of nature (genetics) and nuture (environment). The classical twin study is based on the knowledge that monozygotic (MZ) twins share all of their genes, whereas dizygotic (DZ) twins share on average 50% of the same genes (the same as siblings). Since the early part of the 20th century numerous classical twin studies have been performed to compare concordance rates between identical (MZ) and non-identical (DZ) twins; since the earliest performed twin study in respect to refractive error, conducted in 1922, greater concordance in MZ twin pairs compared to DZ twin pairs has been identified (5). More recent studies have used structural equation modelling to quantify the relative effect of genetics and environment. In this method total variance is estimated by the effect of three factors: the additive genetic effects (A), the dominant (D) genetic or shared environment effects (C), and the unique environmental effects (E), as illustrated in Figure 1.
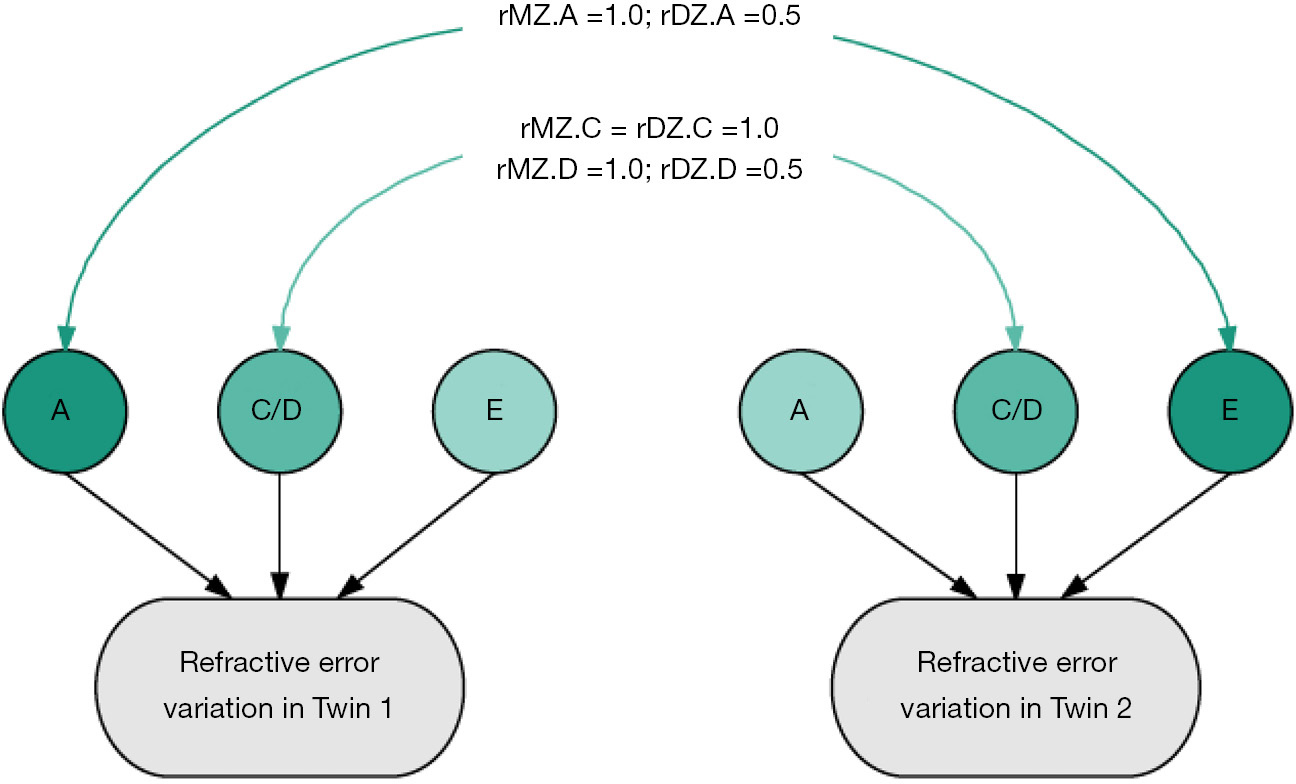
Twin estimates of heritability are consistently high, suggesting genetic factors contribute approximately 70–90% to refractive error variance (9-15). Twin studies have more power to detect heritable effects than family studies (6-8). In 506 British twin pairs from the TwinsUK cohort, univariant twin modeling suggested the variance of refractive error explained by genetic factors was 84–86% (9). This analysis identified the best-fitting model to explain the variance of refractive error was the ‘AE’ model comprising of the additive genetic factors (A) and the unique environmental factors (E). In a subsequent study on an extended TwinsUK cohort (2,301 twin pairs) heritability was estimated from the full ACE model, which included the additional factor of the shared environment (C). The heritability estimate was 77%. This suggests that twin studies have a low power to detect shared environmental factors due to lack of age and generational differences, though these factors may be important (13).
So what do twin studies tell us about genetic risk of myopia and what limitations should be acknowledged? Firstly, twin studies consistently estimate the heritability of refractive error as very high and this has been widely replicated (9-15). Secondly, twin studies enable an estimation of the total contribution of genetic factors—namely they can provide an estimate en masse of the effect of common genetic factors, rare genetic factors, epigenetics, structural genetic factors and to some extent gene-environment interaction factors on trait variance. This is something that cannot currently be estimated using molecular genetic techniques. Thirdly, whilst ascertainment and volunteer bias of twin populations must be considered there is good evidence that rates of disease and health related parameters in twin studies are comparable to singletons within the same population (16). However, twin studies will provide an estimate of heritability that is population specific and therefore, a high heritability estimate may be seen in one population that is not generalizable to another. Reassuringly however heritability estimates from populations around the world are highly comparable (9,12). Additionally heritability estimates are affected by the variation in age of the population when an age-related trait is being studied. This is because in a cohort where there is great variation in age and that is a contributing factor to trait variance, the effect of age will be incorporated into the estimate for the environmental effect (E) and as a result the estimate of heritability will be less. This factor, together with the aforementioned underestimate of shared environmental factors, means that twin studies tend to provide an upper bound estimate of heritability.
The completion of the Human Genome Project in 2003 (17,18) and the subsequent haplotype maps detailing common patterns of genetic variation and inheritance, namely the HapMap project and 1000 Genome Project (19,20), heralded a rush of genome-wide association studies (GWAS) to be performed from 2005 onwards. A GWAS allows a vast number of markers, specifically common genetic polymorphisms (single nucleotide polymorphisms, SNPs), across an individual’s genome to be tested for their association with a disease or trait. The first GWAS to study myopia was performed in 2009 on a cohort with high, pathological myopia. This was followed by a number of similar case-control GWAS of high myopia; although often successful in identifying variants, rates of replication were poor. These and subsequently discussed GWAS are detailed in the GWAS catalog database detailing all published GWAS for myopia, refractive error and other myopia endophenotypes (available at http://www.ebi.ac.uk/gwas/home
Greater success has been obtained when refractive error as a quantitative trait is studied. The first two GWAS for refractive error were published in 2010; both were conducted in European populations and each identified one loci surpassing the GWAS threshold—a loci near the RASGFR1 gene on 15q25.1 and the other near GJD2 on 15q14 (21,22). Whilst subsequent small studies have been performed, the greatest yield by far is obtained by running a GWAS on large datasets. The Consortium for Refractive Error and Myopia (CREAM) is an international collaboration between researchers studying cohorts of both European and Asian descent. A meta-analysis of the GWAS results for refractive error was performed for 37,382 individuals of European descent and 12,332 of Southeast Asian ancestry, published in 2013 (23). The two loci previously identified were replicated and identification of 24 novel loci at genome-wide significance was obtained: BICC1, BMP2, BMP3, CACNA1D, CD55, CHD7, CHRNG, CNDP2, CYP26A1, GJD2, CRIA4, KCNJ2, KCNQ5, LAMA2, MYO1D, PCCA, PRSS56, RASGRF1, RDH5, RORB, SIX6, TOX, ZIC2 and ZMAT4.
At the same time a GWAS performed by the direct-to-consumer genomics company 23andMe (Mountain View, CA, USA) on 55,177 individuals of European descent reported 22 novel loci: BMP3, BMP4, DLG2, DLX1, GJD2, KCNMA1, KCNQ5, LAMA2, LRRC4C, PABPCP2, PDE11A, PRSS56, RASGFR1, RBFOX1, RDH5, RGR, SFRP1, SHISA6, TJP2, TOX, ZBTB38 and ZIC2 (24). Interestingly the authors used the phenotype of self-reported myopia and ‘age of spectacle wear’ as a proxy for severity but obtained remarkably similar results to that obtained in the carefully collected refractive error data in CREAM—virtually all genetic loci were identified in both studies with consistent direction of effect despite analysis on different scales (25,26).
More recently CREAM and 23andMe have combined datasets to perform an even larger GWAS meta-analysis. It is becoming increasingly evident that the only way to capture more of the genetic variation contributing towards to traits is by studying vast numbers of individuals. It is also becoming evident that good proxies of a phenotype, as per the self-reported myopia and age of spectacle wear used by 23andMe, is adequate in genetic association analyses. This metaanalysis comprising over 160,000 individuals is yet to be published but preliminary results have been presented at the Association for Research in Vision and Ophthalmology Meeting; more than 150 loci were identified and association was confirmed for 26 previously reported genes (27,28). Known over-represented functional pathways were confirmed, such as extracellular matrix and ion channel activity, whilst new mechanisms such as angiogenesis, Wnt signalling and TGF-β signalling pathways were suggested. Gene set enrichment analysis to examine important biological pathways strongly supported the importance of light processing as a primary role for the development of refractive error. This recent development in our understanding of functional pathways and mechanisms contributing to myopia risk is fundamental to how genetics may help shape myopia research in the future. The hope is that functional studies in animal models may lead on from genetic studies to more finely examine how genetic polymorphisms translate into myopia development, and in turn highlight how particular steps in how myopia develops that can be targeted therapeutically.
Genetic association studies for refractive error over the last decade have been very successful in identifying loci but how many of the ‘genes for refractive error’ have we identified and what proportion of genetic variance is explained? The answer is sadly that the variance explained by the genetic loci we have identified to date is still rather poor. In the largest published GWAS to date 3.4% of variance is explained by the identified loci (23). The issue of ‘missing heritability’ is well replicated in GWAS of multiple phenotypes such as height and type 2 diabetes (29). We are clearly yet to identify all of the numerous genes of small effect that contribute to myopia risk. Another reason for this missing heritability is GWAS are limited to the contribution of common genetic effects. In a British adolescent cohort the total contribution of common genetic effects on refractive error was estimated at 28%—also known as the SNP based heritability (30). This would suggest that common genetic variants, regardless of whether they have been confirmed to be associated with refractive error, could only ever explain around 30% of the variation of refractive error, just under half of the heritability of estimates of 70–80%. Therefore, the contribution of other genetic effects is likely to be significant and complicated, encompassing rare variants, structural variations, epigenetics and interactions between genes and environment (gene-gene interactions and gene-environment interactions).
A common question posed to genetic researchers is how well does the current genetic knowledge perform at predicting who will develop a trait? Polygenic scores can be used to estimate the variance explained by thousands of genetic variants previously associated (to a varying degree) with the trait of interest. Using the results of the aforementioned meta-analysis between CREAM and 23andMe, the maximum variance explained is estimated to be 7.9% (31). Therefore current polygenic scores can be used to estimate approximately one third of the estimated common genetic heritability—but this obviously falls short of the total estimated heritability and therefore renders the use of genetic prediction in myopia not usable on its own.
However, genetic knowledge may be utilised in different prediction methods, and in the era of emerging treatments for myopia this will become ever more relevant. By combining genetic data, potentially in the form of polygenic risk scores, together with some of the key environmental associations the predictive power of multivariable models may be increased. The age of spectacle wear in particular is highly useful in predicating the final severity of myopia in adulthood, however this factor will obviously post-date the period in which risk of myopia developing myopia would be assessed. In the CLEERE study thirteen predictive factors of incident myopia were assessed—the single best risk factor was refractive error at baseline (4). The authors attempted to incorporate genetic risk using the number of myopic parents as an additional factor in their predictive model—this conferred no significant benefit over the use of refractive error at baseline alone. However, as Guggenheim et al argue in a recent review, parental myopia is only a good predictor of offspring’s risk of myopia on average and therefore unlikely to predict which particular child will fall into the extremes of refractive error (32).
The concept of ‘personalized medicine’ with tailored treatment to individuals has received much clinical interest—in terms of myopia the hope is that we may identify genotypes that respond better to certain treatments, or environmental factors that interact to either increase or reduce genetic susceptibility to myopia. Whilst, for example, a genotype that responds acutely well to atropine therapy is yet to be identified, there is evidence that a specific myopia associated allele interacts with time spent reading to increase myopia risk (33). Gene-environment interactions have yet to be identified for any other myopia associations—of note no interaction between genes and the highly protective effect of time outdoors has been identified. The inference being that to date gene-environment interactions cannot play a role in myopia prediction, however future research may yet highlight genotypes that may play a role in personalized medicine.
There are huge differences in the rates of myopia between Asian and Western Countries. The reasons for this are multifactorial with environmental pressures playing a key role but what evidence is there that genetics play a role? To some extent this question cannot be fully answered. The majority of large GWAS in refractive error have been performed in predominantly European populations—the aforementioned 23andMe dataset is purely of European descent whilst the CREAM dataset is predominantly European with approximately one quarter of Asian descent. Also, many of the heritability estimates are based on European populations. This is very relevant to myopia prediction as both genetic prediction scores and heritability estimates will be specific to the population (and ethnicity) in which they were identified. However, what has been identified from the published CREAM meta-analysis, is that whilst many of the loci that were significant in the European cohorts were non-significant in the smaller Asian contributing studies, due to low power, the loci mostly had a similar effect size and direction as in the European ancestry sample (23). The research pointed towards evidence for shared genetic risk rather differing genetic risk, and therefore would suggest differences in prevalence rates are not due to genetics. This is conferred by the fact that heritability estimates from Asian populations are largely comparable to European populations (14,15). One must also acknowledge that if the Asian myopia epidemic was due to genetic factors, the time in which genetic evolution could be attributable would be far greater than the dramatic rises we have seen over approximately the last 50 years.
Myopia is a highly heritable trait and genetics undoubtedly play a significant role in myopia risk, although they have not led to the recent myopia epidemic. Identifying all the ways in which genetic risk is conferred is vast and complicated. Whilst genetic research techniques develop rapidly, the main achievable target for myopia researchers is reaching a point whereby a measurable and relevant level of common genetic risk factors can be assessed and incorporated into risk prediction models. Specific risk genotypes that can tailor treatment or lifestyle advice are yet to be identified but may also play a role in the near future. The incorporation of this information into the targeted treatment of individuals at risk of myopia, or perhaps just high myopia, is the proximal target for researchers whilst longer term the hope is that a greater understanding of myopia genetics and developmental mechanisms will help to reduce the myopia burden of the future.