1、Jager MJ, Shields CL, Cebulla CM, et al. Uveal melanoma[ J]. Nat Rev Dis Primers, 2020, 6(1): 24.Jager MJ, Shields CL, Cebulla CM, et al. Uveal melanoma[ J]. Nat Rev Dis Primers, 2020, 6(1): 24.
2、Smit KN, Jager MJ, de Klein A, et al. Uveal melanoma: towards a molecular understanding[ J]. Prog Retin Eye Res, 2020, 75: 100800.Smit KN, Jager MJ, de Klein A, et al. Uveal melanoma: towards a molecular understanding[ J]. Prog Retin Eye Res, 2020, 75: 100800.
3、Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma[ J]. Cancer Cell,2017, 32(2): 204-220.e15.Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma[ J]. Cancer Cell,2017, 32(2): 204-220.e15.
4、Ransohoff JD, Wei Y, Khavari PA, et al. The functions and unique features of long intergenic non-coding RNA[ J]. Nat Rev Mol Cell Biol,2018, 19(3): 143-157.Ransohoff JD, Wei Y, Khavari PA, et al. The functions and unique features of long intergenic non-coding RNA[ J]. Nat Rev Mol Cell Biol,2018, 19(3): 143-157.
5、Schmitt AM, Chang HY, et al. Long noncoding RNAs in cancer pathways[ J]. Cancer Cell, 2016, 29(4): 452-463.Schmitt AM, Chang HY, et al. Long noncoding RNAs in cancer pathways[ J]. Cancer Cell, 2016, 29(4): 452-463.
6、Tay Y, Rinn J, Pandolfi PP, et al. The multilayered complexity of ceRNA crosstalk and competition[ J]. Nature, 2014, 505(7483): 344-352.Tay Y, Rinn J, Pandolfi PP, et al. The multilayered complexity of ceRNA crosstalk and competition[ J]. Nature, 2014, 505(7483): 344-352.
7、Chen X, Chen Z, Yu S, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer[ J]. Clin Cancer Res, 2018,24(8): 2002-2014.Chen X, Chen Z, Yu S, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer[ J]. Clin Cancer Res, 2018,24(8): 2002-2014.
8、Li H, Wang X, Wen C, et al. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer[ J].Mol Cancer, 2017, 16(1): 169.Li H, Wang X, Wen C, et al. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer[ J].Mol Cancer, 2017, 16(1): 169.
9、Lu Q, Zhao N, Zha G, et al. LncRNA HOXA11-AS exerts oncogenic functions by repressing p21 and miR-124 in uveal melanoma[ J]. DNA Cell Biol, 2017, 36(10): 837-844.Lu Q, Zhao N, Zha G, et al. LncRNA HOXA11-AS exerts oncogenic functions by repressing p21 and miR-124 in uveal melanoma[ J]. DNA Cell Biol, 2017, 36(10): 837-844.
10、Lu L, Yu X, Zhang L, et al. The long non-coding RNA RHPN1-AS1 promotes uveal melanoma progression[J]. Int J Mol Sci, 2017, 18(1): 226.Lu L, Yu X, Zhang L, et al. The long non-coding RNA RHPN1-AS1 promotes uveal melanoma progression[J]. Int J Mol Sci, 2017, 18(1): 226.
11、Xu H, Gong J, Liu H, et al. High expression of lncRNA PVT1 independently predicts poor overall survival in patients with primary uveal melanoma[ J]. PLoS One, 2017, 12(12): e0189675.Xu H, Gong J, Liu H, et al. High expression of lncRNA PVT1 independently predicts poor overall survival in patients with primary uveal melanoma[ J]. PLoS One, 2017, 12(12): e0189675.
12、Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion[ J]. Nat Commun, 2017, 8(1): 289.Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion[ J]. Nat Commun, 2017, 8(1): 289.
13、Liang H, Yu T, Han Y, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression[ J]. Mol Cancer, 2018, 17(1): 119.Liang H, Yu T, Han Y, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression[ J]. Mol Cancer, 2018, 17(1): 119.
14、Li Y, Zeng C, Hu J, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression[ J]. J Hematol Oncol,2018, 11(1): 89.Li Y, Zeng C, Hu J, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression[ J]. J Hematol Oncol,2018, 11(1): 89.
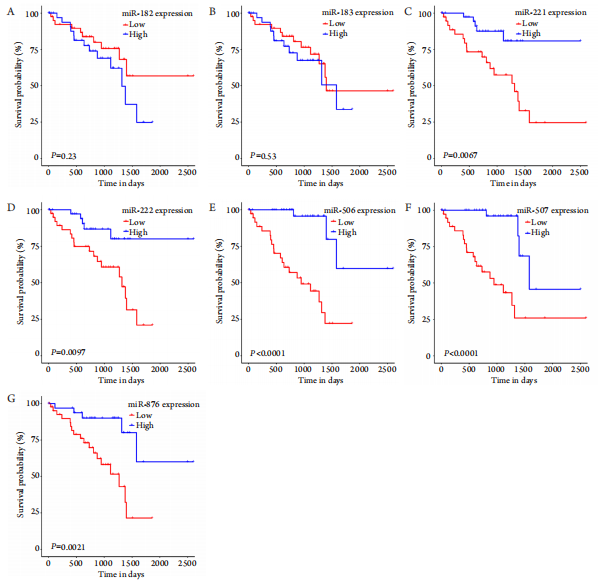
15、Jia L, Liu W, Cao B, et al. MiR-507 inhibits the migration and invasion of human breastcancer cells through Flt-1 suppression[ J]. Oncotarget,2016, 7(24): 36743-36754.Jia L, Liu W, Cao B, et al. MiR-507 inhibits the migration and invasion of human breastcancer cells through Flt-1 suppression[ J]. Oncotarget,2016, 7(24): 36743-36754.
16、Wei Y, Sun Q, Zhao L, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma[ J]. Med Oncol, 2016, 33(8): 88.Wei Y, Sun Q, Zhao L, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma[ J]. Med Oncol, 2016, 33(8): 88.
17、Dong Y, Zheng Y, Wang C, et al. MiR-876-5p modulates head and neck squamous cell carcinoma metastasis and invasion by targeting vimentin[ J]. Cancer Cell Int, 2018, 18: 121.Dong Y, Zheng Y, Wang C, et al. MiR-876-5p modulates head and neck squamous cell carcinoma metastasis and invasion by targeting vimentin[ J]. Cancer Cell Int, 2018, 18: 121.
18、Fornari F, Pollutri D, Patrizi C, et al. In hepatocellular carcinoma miR-221 modulates sorafenib resistance through inhibition of caspase-3-mediated apoptosis[ J]. Clin Cancer Res, 2017, 23(14): 3953-3965.Fornari F, Pollutri D, Patrizi C, et al. In hepatocellular carcinoma miR-221 modulates sorafenib resistance through inhibition of caspase-3-mediated apoptosis[ J]. Clin Cancer Res, 2017, 23(14): 3953-3965.
19、Zhou CF, Ma J, Huang L, et al. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1[J]. Oncogene, 2019, 38(8): 1256-1268.Zhou CF, Ma J, Huang L, et al. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1[J]. Oncogene, 2019, 38(8): 1256-1268.
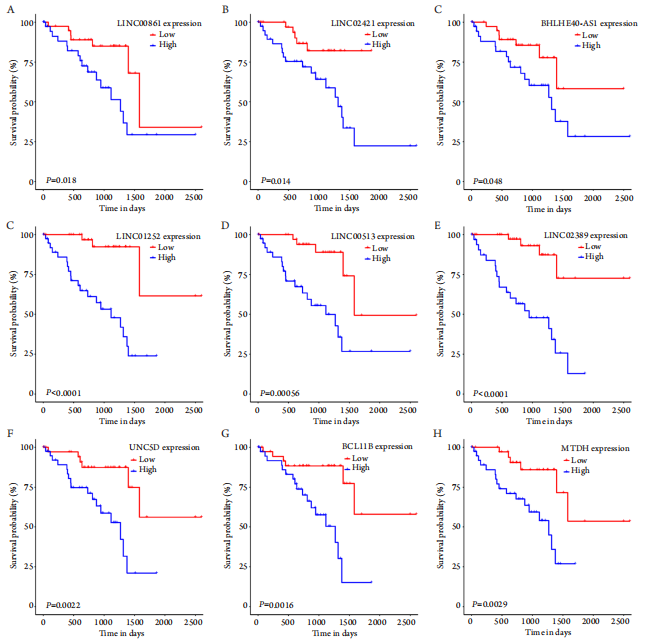
20、DeVaux RS, Ropri AS, Grimm SL, et al. Long noncoding RNA BHLHE40-AS1 promotes early breast cancer progression through modulating IL-6/STAT3 signaling[J]. J Cell Biochem, 2020, 121(7): 3465-3478.DeVaux RS, Ropri AS, Grimm SL, et al. Long noncoding RNA BHLHE40-AS1 promotes early breast cancer progression through modulating IL-6/STAT3 signaling[J]. J Cell Biochem, 2020, 121(7): 3465-3478.
21、Lv Y, Wei W, Huang Z, et al. Long non-coding RNA expression profile can predict early recurrence in hepatocellular carcinoma after curative resection[ J]. Hepatol Res, 2018, 48(13): 1140-1148.Lv Y, Wei W, Huang Z, et al. Long non-coding RNA expression profile can predict early recurrence in hepatocellular carcinoma after curative resection[ J]. Hepatol Res, 2018, 48(13): 1140-1148.
22、Zhu B, Chen H, Zhang X, et al. Serum miR-30d as a novel biomarker for multiple myeloma and its antitumor role in U266 cells through the targeting of the MTDH/PI3K/Akt signaling pathway[ J]. Int J Oncol,2018, 53(5): 2131-2144.Zhu B, Chen H, Zhang X, et al. Serum miR-30d as a novel biomarker for multiple myeloma and its antitumor role in U266 cells through the targeting of the MTDH/PI3K/Akt signaling pathway[ J]. Int J Oncol,2018, 53(5): 2131-2144.
23、Yu DP, Zhou Y. Astrocyte elevated gene 1 (AEG-1) acts as a promoter gene in clear cell renal cell carcinoma cell growth and metastasis[ J].Med Sci Monit, 2018, 24: 8213-8223.Yu DP, Zhou Y. Astrocyte elevated gene 1 (AEG-1) acts as a promoter gene in clear cell renal cell carcinoma cell growth and metastasis[ J].Med Sci Monit, 2018, 24: 8213-8223.
24、Li JW, Huang CZ, Li JH, et al. Knockdown of metadherin inhibits cell proliferation and migration in colorectal cancer[ J]. Oncol Rep, 2018,40(4): 2215-2223.Li JW, Huang CZ, Li JH, et al. Knockdown of metadherin inhibits cell proliferation and migration in colorectal cancer[ J]. Oncol Rep, 2018,40(4): 2215-2223.
25、Zhang Y, Peng G, Wang Y, et al. Silencing of astrocyte elevated gene-1 inhibits proliferation and migration of melanoma cells and induces apoptosis[ J]. Clin Exp Pharmacol Physiol, 2017, 44(7): 815-826.Zhang Y, Peng G, Wang Y, et al. Silencing of astrocyte elevated gene-1 inhibits proliferation and migration of melanoma cells and induces apoptosis[ J]. Clin Exp Pharmacol Physiol, 2017, 44(7): 815-826.
26、Kraszewska MD, Dawidowska M, Kosmalska M, et al. BCL11B, FLT3,NOTCH1 and FBXW7 mutation status in T-cell acute lymphoblastic leukemia patients[ J]. Blood Cells Mol Dis, 2013, 50(1): 33-38.Kraszewska MD, Dawidowska M, Kosmalska M, et al. BCL11B, FLT3,NOTCH1 and FBXW7 mutation status in T-cell acute lymphoblastic leukemia patients[ J]. Blood Cells Mol Dis, 2013, 50(1): 33-38.
27、Liao CK, Fang KM, Chai K, et al. Depletion of B cell CLL/lymphoma 11B gene expression represses glioma cell growth[ J]. Mol Neurobiol,2016, 53(6): 3528-3539.Liao CK, Fang KM, Chai K, et al. Depletion of B cell CLL/lymphoma 11B gene expression represses glioma cell growth[ J]. Mol Neurobiol,2016, 53(6): 3528-3539.