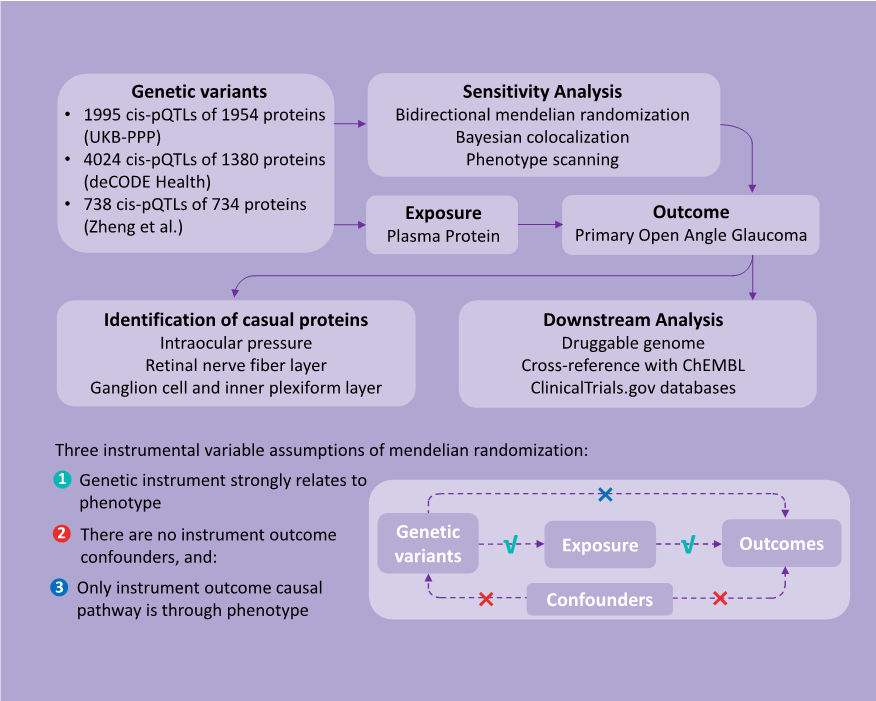
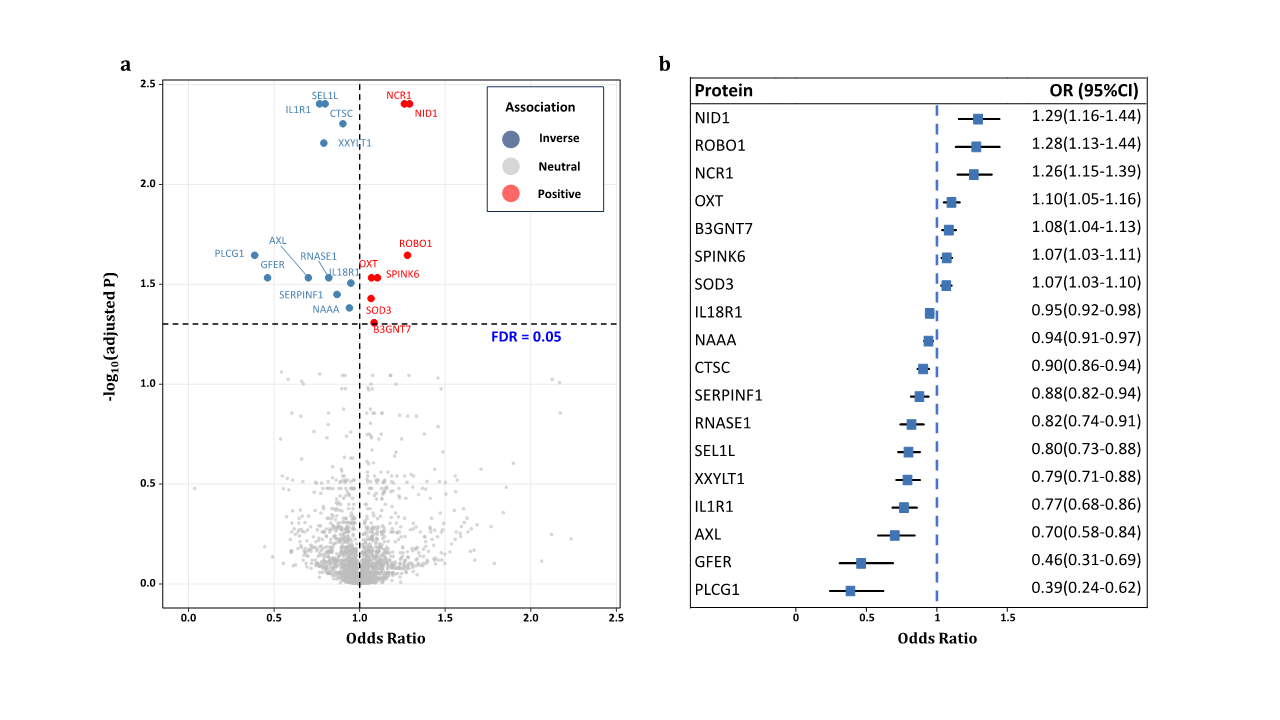
1、Stein JD, Khawaja AP, Weizer JS. Glaucoma in adults�screening, diagnosis, and management: a review. JAMA.
2021, 325(2): 164-174. DOI: 10.1001/jama.2020.21899.Stein JD, Khawaja AP, Weizer JS. Glaucoma in adults�screening, diagnosis, and management: a review. JAMA.
2021, 325(2): 164-174. DOI: 10.1001/jama.2020.21899.
2、Tham YC, Li X, Wong TY, et al. Global prevalence
of glaucoma and projections of glaucoma burden
through 2040: a systematic review and meta-analysis.
Ophthalmology. 2014, 121(11): 2081-2090. DOI: 10.1016/
j.ophtha.2014.05.013.Tham YC, Li X, Wong TY, et al. Global prevalence
of glaucoma and projections of glaucoma burden
through 2040: a systematic review and meta-analysis.
Ophthalmology. 2014, 121(11): 2081-2090. DOI: 10.1016/
j.ophtha.2014.05.013.
3、Gedde%20SJ%2C%20Vinod%20K%2C%20Wright%20MM%2C%20et%20al.%20Primary%20%0Aopen-angle%20glaucoma%20preferred%20practice%20pattern%C2%AE.%20%0AOphthalmology.%202021%2C%20128(1)%3A%20P71-P150.%20DOI%3A%2010.1016%2F%0Aj.ophtha.2020.10.022.Gedde%20SJ%2C%20Vinod%20K%2C%20Wright%20MM%2C%20et%20al.%20Primary%20%0Aopen-angle%20glaucoma%20preferred%20practice%20pattern%C2%AE.%20%0AOphthalmology.%202021%2C%20128(1)%3A%20P71-P150.%20DOI%3A%2010.1016%2F%0Aj.ophtha.2020.10.022.
4、Jayaram H, Kolko M, Friedman DS, et al. Glaucoma: now
and beyond. Lancet. 2023, 402(10414): 1788-1801. DOI:
10.1016/S0140-6736(23)01289-8.Jayaram H, Kolko M, Friedman DS, et al. Glaucoma: now
and beyond. Lancet. 2023, 402(10414): 1788-1801. DOI:
10.1016/S0140-6736(23)01289-8.
5、 Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory
networks of human serum proteins link genetics to disease.
Science. 2018, 361(6404): 769-773. DOI: 10.1126/science.
aaq1327. Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory
networks of human serum proteins link genetics to disease.
Science. 2018, 361(6404): 769-773. DOI: 10.1126/science.
aaq1327.
6、 Xu JY, Zhang C, Wang X, et al. Integrative proteomic
characterization of human lung adenocarcinoma.
Cell. 2020, 182(1): 245-261.e17. DOI: 10.1016/j.cell.2020.05.043. Xu JY, Zhang C, Wang X, et al. Integrative proteomic
characterization of human lung adenocarcinoma.
Cell. 2020, 182(1): 245-261.e17. DOI: 10.1016/j.cell.2020.05.043.
7、Chouliaras L, Thomas A, Malpetti M, et al. Differential
levels of plasma biomarkers of neurodegeneration in
Lewy body dementia, Alzheimer’s disease, frontotemporal
dementia and progressive supranuclear palsy. J Neurol
Neurosurg Psychiatry. 2022, 93(6): 651-658. DOI:
10.1136/jnnp-2021-327788.Chouliaras L, Thomas A, Malpetti M, et al. Differential
levels of plasma biomarkers of neurodegeneration in
Lewy body dementia, Alzheimer’s disease, frontotemporal
dementia and progressive supranuclear palsy. J Neurol
Neurosurg Psychiatry. 2022, 93(6): 651-658. DOI:
10.1136/jnnp-2021-327788.
8、Chan MY, Efthymios M, Tan SH, et al. Prioritizing
candidates of post-myocardial infarction heart failure
using plasma proteomics and single-cell transcriptomics.
Circulation. 2020, 142(15): 1408-1421. DOI: 10.1161/
CIRCULATIONAHA.119.045158.Chan MY, Efthymios M, Tan SH, et al. Prioritizing
candidates of post-myocardial infarction heart failure
using plasma proteomics and single-cell transcriptomics.
Circulation. 2020, 142(15): 1408-1421. DOI: 10.1161/
CIRCULATIONAHA.119.045158.
9、Santos R, Ursu O, Gaulton A, et al. A comprehensive map
of molecular drug targets. Nat Rev Drug Discov. 2017,
16(1): 19-34. DOI: 10.1038/nrd.2016.230.Santos R, Ursu O, Gaulton A, et al. A comprehensive map
of molecular drug targets. Nat Rev Drug Discov. 2017,
16(1): 19-34. DOI: 10.1038/nrd.2016.230.
10、Davey Smith G, Hemani G. Mendelian randomization:
genetic anchors for causal inference in epidemiological
studies. Hum Mol Genet. 2014, 23(R1): R89-R98. DOI:
10.1093/hmg/ddu328.Davey Smith G, Hemani G. Mendelian randomization:
genetic anchors for causal inference in epidemiological
studies. Hum Mol Genet. 2014, 23(R1): R89-R98. DOI:
10.1093/hmg/ddu328.
11、 Nusinovici S, Li H, Thakur S, et al. High-density
lipoprotein 3 cholesterol and primary open-angle
glaucoma: metabolomics and Mendelian randomization
analyses. Ophthalmology. 2022, 129(3): 285-294. DOI:
10.1016/j.ophtha.2021.09.013. Nusinovici S, Li H, Thakur S, et al. High-density
lipoprotein 3 cholesterol and primary open-angle
glaucoma: metabolomics and Mendelian randomization
analyses. Ophthalmology. 2022, 129(3): 285-294. DOI:
10.1016/j.ophtha.2021.09.013.
12、Hu Z, Zhou F, Kaminga AC, et al. Type 2 diabetes, fasting
glucose, hemoglobin A1c levels and risk of primary open�angle glaucoma: a Mendelian randomization study. Invest
Ophthalmol Vis Sci. 2022, 63(5): 37. DOI: 10.1167/
iovs.63.5.37.Hu Z, Zhou F, Kaminga AC, et al. Type 2 diabetes, fasting
glucose, hemoglobin A1c levels and risk of primary open�angle glaucoma: a Mendelian randomization study. Invest
Ophthalmol Vis Sci. 2022, 63(5): 37. DOI: 10.1167/
iovs.63.5.37.
13、Rajasundaram S, Zebardast N, Mehta P, et al. TIE1 and
TEK signalling, intraocular pressure, and primary open�angle glaucoma: a Mendelian randomization study. J
Transl Med. 2023, 21(1): 847. DOI: 10.1186/s12967-023-
04737-9.Rajasundaram S, Zebardast N, Mehta P, et al. TIE1 and
TEK signalling, intraocular pressure, and primary open�angle glaucoma: a Mendelian randomization study. J
Transl Med. 2023, 21(1): 847. DOI: 10.1186/s12967-023-
04737-9.
14、Pietzner M, Wheeler E, Carrasco-Zanini J, et al. Mapping
the proteo-genomic convergence of human diseases.
Science. 2021, 374(6569): eabj1541. DOI: 10.1126/science.abj1541.Pietzner M, Wheeler E, Carrasco-Zanini J, et al. Mapping
the proteo-genomic convergence of human diseases.
Science. 2021, 374(6569): eabj1541. DOI: 10.1126/science.abj1541.
15、Ferkingstad E, Sulem P, Atlason BA, et al. Large-scale
integration of the plasma proteome with genetics and
disease. Nat Genet. 2021, 53(12): 1712-1721. DOI:
10.1038/s41588-021-00978-w.Ferkingstad E, Sulem P, Atlason BA, et al. Large-scale
integration of the plasma proteome with genetics and
disease. Nat Genet. 2021, 53(12): 1712-1721. DOI:
10.1038/s41588-021-00978-w.
16、Ursu O, Glick M, Oprea T. Novel drug targets in 2018.
Nat Rev Drug Discov. 2019. DOI: 10.1038/d41573-019-
00052-5.Ursu O, Glick M, Oprea T. Novel drug targets in 2018.
Nat Rev Drug Discov. 2019. DOI: 10.1038/d41573-019-
00052-5.
17、Yao C, Chen G, Song C, et al. Genome-wide mapping of
plasma protein QTLs identifies putatively causal genes and
pathways for cardiovascular disease. Nat Commun. 2018,
9(1): 3268. DOI: 10.1038/s41467-018-05512-x.Yao C, Chen G, Song C, et al. Genome-wide mapping of
plasma protein QTLs identifies putatively causal genes and
pathways for cardiovascular disease. Nat Commun. 2018,
9(1): 3268. DOI: 10.1038/s41467-018-05512-x.
18、Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of
the human plasma proteome. Nature. 2018, 558(7708): 73-
79. DOI: 10.1038/s41586-018-0175-2.Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of
the human plasma proteome. Nature. 2018, 558(7708): 73-
79. DOI: 10.1038/s41586-018-0175-2.
19、Hulur I, Gamazon ER, Skol AD, et al. Enrichment of
inflammatory bowel disease and colorectal cancer risk
variants in colon expression quantitative trait loci. BMC
Genomics. 2015, 16(1): 138. DOI: 10.1186/s12864-015-
1292-z.Hulur I, Gamazon ER, Skol AD, et al. Enrichment of
inflammatory bowel disease and colorectal cancer risk
variants in colon expression quantitative trait loci. BMC
Genomics. 2015, 16(1): 138. DOI: 10.1186/s12864-015-
1292-z.
20、 Sun BB, Chiou J, Traylor M, et al. Plasma proteomic
associations with genetics and health in the UK Biobank.
Nature. 2023, 622(7982): 329-338. DOI: 10.1038/s41586-
023-06592-6. Sun BB, Chiou J, Traylor M, et al. Plasma proteomic
associations with genetics and health in the UK Biobank.
Nature. 2023, 622(7982): 329-338. DOI: 10.1038/s41586-
023-06592-6.
21、Zheng J, Haberland V, Baird D, et al. Phenome-wide
Mendelian randomization mapping the influence of the
plasma proteome on complex diseases. Nat Genet. 2020,
52(10): 1122-1131. DOI: 10.1038/s41588-020-0682-6.Zheng J, Haberland V, Baird D, et al. Phenome-wide
Mendelian randomization mapping the influence of the
plasma proteome on complex diseases. Nat Genet. 2020,
52(10): 1122-1131. DOI: 10.1038/s41588-020-0682-6.
22、Kurki MI, Karjalainen J, Palta P, et al. FinnGen provides
genetic insights from a well-phenotyped isolated
population. Nature. 2023, 613(7944): 508-518. DOI:
10.1038/s41586-022-05473-8.Kurki MI, Karjalainen J, Palta P, et al. FinnGen provides
genetic insights from a well-phenotyped isolated
population. Nature. 2023, 613(7944): 508-518. DOI:
10.1038/s41586-022-05473-8.
23、Springelkamp H, Iglesias AI, Cuellar-Partida G, et al.
ARHGEF12 influences the risk of glaucoma by increasing
intraocular pressure. Hum Mol Genet. 2015, 24(9): 2689-
2699. DOI: 10.1093/hmg/ddv027.Springelkamp H, Iglesias AI, Cuellar-Partida G, et al.
ARHGEF12 influences the risk of glaucoma by increasing
intraocular pressure. Hum Mol Genet. 2015, 24(9): 2689-
2699. DOI: 10.1093/hmg/ddv027.
24、Currant H, Hysi P, Fitzgerald TW, et al. Genetic variation
affects morphological retinal phenotypes extracted from
UK Biobank optical coherence tomography images. PLoS
Genet. 2021, 17(5): e1009497. DOI: 10.1371/journal.
pgen.1009497.Currant H, Hysi P, Fitzgerald TW, et al. Genetic variation
affects morphological retinal phenotypes extracted from
UK Biobank optical coherence tomography images. PLoS
Genet. 2021, 17(5): e1009497. DOI: 10.1371/journal.
pgen.1009497.
25、Pierce BL, Burgess S. Efficient design for Mendelian
randomization studies: subsample and 2-sample
instrumental variable estimators. Am J Epidemiol. 2013,
178(7): 1177-1184. DOI: 10.1093/aje/kwt084.Pierce BL, Burgess S. Efficient design for Mendelian
randomization studies: subsample and 2-sample
instrumental variable estimators. Am J Epidemiol. 2013,
178(7): 1177-1184. DOI: 10.1093/aje/kwt084.
26、Foley CN, Staley JR, Breen PG, et al. A fast and efficient
colocalization algorithm for identifying shared genetic risk
factors across multiple traits. Nat Commun. 2021, 12(1):
764. DOI: 10.1038/s41467-020-20885-8.Foley CN, Staley JR, Breen PG, et al. A fast and efficient
colocalization algorithm for identifying shared genetic risk
factors across multiple traits. Nat Commun. 2021, 12(1):
764. DOI: 10.1038/s41467-020-20885-8.
27、Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner
V2: an expanded tool for searching human genotype�phenotype associations. Bioinformatics. 2019, 35(22):
4851-4853. DOI: 10.1093/bioinformatics/btz469.Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner
V2: an expanded tool for searching human genotype�phenotype associations. Bioinformatics. 2019, 35(22):
4851-4853. DOI: 10.1093/bioinformatics/btz469.
28、Finan C, Gaulton A, Kruger FA, et al. The druggable
genome and support for target identification and validation
in drug development. Sci Transl Med. 2017, 9(383):
eaag1166. DOI: 10.1126/scitranslmed.aag1166.Finan C, Gaulton A, Kruger FA, et al. The druggable
genome and support for target identification and validation
in drug development. Sci Transl Med. 2017, 9(383):
eaag1166. DOI: 10.1126/scitranslmed.aag1166.
29、 Mendez D, Gaulton A, Bento AP, et al. ChEMBL: towards
direct deposition of bioassay data. Nucleic Acids Res.
2019, 47(D1): D930-D940. DOI: 10.1093/nar/gky1075. Mendez D, Gaulton A, Bento AP, et al. ChEMBL: towards
direct deposition of bioassay data. Nucleic Acids Res.
2019, 47(D1): D930-D940. DOI: 10.1093/nar/gky1075.
30、Bhattacharya A, Wei J, Song W, et al. SEL1L-HRD1
ER-associated degradation suppresses hepatocyte
hyperproliferation and liver cancer. iScience. 2022, 25(10):
105183. DOI: 10.1016/j.isci.2022.105183.Bhattacharya A, Wei J, Song W, et al. SEL1L-HRD1
ER-associated degradation suppresses hepatocyte
hyperproliferation and liver cancer. iScience. 2022, 25(10):
105183. DOI: 10.1016/j.isci.2022.105183.
31、Hattori T, Hanafusa K, Wada I, et al. SEL1L degradation
intermediates stimulate cytosolic aggregation of
polyglutamine-expanded protein. FEBS J. 2021, 288(15):
4637-4654. DOI: 10.1111/febs.15761.Hattori T, Hanafusa K, Wada I, et al. SEL1L degradation
intermediates stimulate cytosolic aggregation of
polyglutamine-expanded protein. FEBS J. 2021, 288(15):
4637-4654. DOI: 10.1111/febs.15761.
32、Hwang J, Qi L. Quality Control in the Endoplasmic
Reticulum: Crosstalk between ERAD and UPR pathways.
Trends Biochem Sci. 2018, 43(8): 593-605. DOI: 10.1016/
j.tibs.2018.06.005.Hwang J, Qi L. Quality Control in the Endoplasmic
Reticulum: Crosstalk between ERAD and UPR pathways.
Trends Biochem Sci. 2018, 43(8): 593-605. DOI: 10.1016/
j.tibs.2018.06.005.
33、Bhosle VK, Tan JM, Li T, et al. SLIT2/ROBO1 signaling
suppresses mTORC1 for organelle control and bacterial
killing. Life Sci Alliance. 2023, 6(8): e202301964. DOI:
10.26508/lsa.202301964.Bhosle VK, Tan JM, Li T, et al. SLIT2/ROBO1 signaling
suppresses mTORC1 for organelle control and bacterial
killing. Life Sci Alliance. 2023, 6(8): e202301964. DOI:
10.26508/lsa.202301964.
34、Blockus H, Chédotal A. Slit-robo signaling. Development.
2016, 143(17): 3037-3044. DOI: 10.1242/dev.132829.Blockus H, Chédotal A. Slit-robo signaling. Development.
2016, 143(17): 3037-3044. DOI: 10.1242/dev.132829.
35、Courtney JM, Ye Q, Nesbitt AE, et al. Experimental
protein structure verification by scoring with a single,
unassigned NMR spectrum. Structure. 2015, 23(10): 1958-
1966. DOI: 10.1016/j.str.2015.07.019.Courtney JM, Ye Q, Nesbitt AE, et al. Experimental
protein structure verification by scoring with a single,
unassigned NMR spectrum. Structure. 2015, 23(10): 1958-
1966. DOI: 10.1016/j.str.2015.07.019.
36、Gan YJ, Cao Y, Zhang ZH, et al. Srgap2 suppression
ameliorates retinal ganglion cell degeneration in mice.
Neural Regen Res. 2023, 18(10): 2307-2314. DOI:
10.4103/1673-5374.369122.Gan YJ, Cao Y, Zhang ZH, et al. Srgap2 suppression
ameliorates retinal ganglion cell degeneration in mice.
Neural Regen Res. 2023, 18(10): 2307-2314. DOI:
10.4103/1673-5374.369122.
37、Huang L, Xu Y, Yu W, et al. Effect of Robo1 on retinal
pigment epithelial cells and experimental proliferative
vitreoretinopathy. Invest Ophthalmol Vis Sci. 2010, 51(6):
3193-3204. DOI: 10.1167/iovs.09-3779.Huang L, Xu Y, Yu W, et al. Effect of Robo1 on retinal
pigment epithelial cells and experimental proliferative
vitreoretinopathy. Invest Ophthalmol Vis Sci. 2010, 51(6):
3193-3204. DOI: 10.1167/iovs.09-3779.
38、Rama N, Dubrac A, Mathivet T, et al. Slit2 signaling
through Robo1 and Robo2 is required for retinal
neovascularization. Nat Med. 2015, 21(5): 483-491. DOI:
10.1038/nm.3849.Rama N, Dubrac A, Mathivet T, et al. Slit2 signaling
through Robo1 and Robo2 is required for retinal
neovascularization. Nat Med. 2015, 21(5): 483-491. DOI:
10.1038/nm.3849.
39、Balasubramani M, Schreiber EM, Candiello J, et al.
Molecular interactions in the retinal basement membrane
system: a proteomic approach. Matrix Biol. 2010, 29(6):
471-483. DOI: 10.1016/j.matbio.2010.04.002.Balasubramani M, Schreiber EM, Candiello J, et al.
Molecular interactions in the retinal basement membrane
system: a proteomic approach. Matrix Biol. 2010, 29(6):
471-483. DOI: 10.1016/j.matbio.2010.04.002.
40、Jayadev R, Sherwood DR. Basement membranes. Curr
Biol. 2017, 27(6): R207-R211. DOI: 10.1016/j.cub.
2017.02.006.Jayadev R, Sherwood DR. Basement membranes. Curr
Biol. 2017, 27(6): R207-R211. DOI: 10.1016/j.cub.
2017.02.006.
41、Vranka JA, Kelley MJ, Acott TS, et al. Extracellular matrix
in the trabecular meshwork: intraocular pressure regulation
and dysregulation in glaucoma. Exp Eye Res. 2015, 133:
112-125. DOI: 10.1016/j.exer.2014.07.014.Vranka JA, Kelley MJ, Acott TS, et al. Extracellular matrix
in the trabecular meshwork: intraocular pressure regulation
and dysregulation in glaucoma. Exp Eye Res. 2015, 133:
112-125. DOI: 10.1016/j.exer.2014.07.014.
42、Ibrahim S, Weiss TS. Augmenter of liver regeneration:
essential for growth and beyond. Cytokine Growth Factor
Rev. 2019, 45: 65-80. DOI: 10.1016/j.cytogfr.2018.12.003.Ibrahim S, Weiss TS. Augmenter of liver regeneration:
essential for growth and beyond. Cytokine Growth Factor
Rev. 2019, 45: 65-80. DOI: 10.1016/j.cytogfr.2018.12.003.
43、Verma AK, Sharma A, Subramaniyam N, et al. Augmenter
of liver regeneration: Mitochondrial function and
steatohepatitis. J Hepatol. 2022, 77(5): 1410-1421. DOI: 10.1016/j.jhep.2022.06.019.Verma AK, Sharma A, Subramaniyam N, et al. Augmenter
of liver regeneration: Mitochondrial function and
steatohepatitis. J Hepatol. 2022, 77(5): 1410-1421. DOI: 10.1016/j.jhep.2022.06.019.
44、Weinreb RN, Aung T, Medeiros FA. The pathophysiology
and treatment of glaucoma: a review. JAMA. 2014,
311(18): 1901-1911. DOI: 10.1001/jama.2014.3192.Weinreb RN, Aung T, Medeiros FA. The pathophysiology
and treatment of glaucoma: a review. JAMA. 2014,
311(18): 1901-1911. DOI: 10.1001/jama.2014.3192.
45、Ju WK, Perkins GA, Kim KY, et al. Glaucomatous optic
neuropathy: Mitochondrial dynamics, dysfunction and
protection in retinal ganglion cells. Prog Retin Eye Res.
2023, 95: 101136. DOI: 10.1016/j.preteyeres.2022.101136.Ju WK, Perkins GA, Kim KY, et al. Glaucomatous optic
neuropathy: Mitochondrial dynamics, dysfunction and
protection in retinal ganglion cells. Prog Retin Eye Res.
2023, 95: 101136. DOI: 10.1016/j.preteyeres.2022.101136.
46、Zhu C, Wei Y, Wei X. AXL receptor tyrosine kinase as
a promising anti-cancer approach: functions, molecular
mechanisms and clinical applications. Mol Cancer. 2019,
18(1): 153. DOI: 10.1186/s12943-019-1090-3.Zhu C, Wei Y, Wei X. AXL receptor tyrosine kinase as
a promising anti-cancer approach: functions, molecular
mechanisms and clinical applications. Mol Cancer. 2019,
18(1): 153. DOI: 10.1186/s12943-019-1090-3.
47、Zhao W, Fan J, Kulic I, et al. Axl receptor tyrosine kinase
is a regulator of apolipoprotein E. Mol Brain. 2020, 13(1):
66. DOI: 10.1186/s13041-020-00609-1.Zhao W, Fan J, Kulic I, et al. Axl receptor tyrosine kinase
is a regulator of apolipoprotein E. Mol Brain. 2020, 13(1):
66. DOI: 10.1186/s13041-020-00609-1.
48、Margeta MA, Yin Z, Madore C, et al. Apolipoprotein
E4 impairs the response of neurodegenerative retinal
microglia and prevents neuronal loss in glaucoma.
Immunity. 2022, 55(9): 1627-1644.e7. DOI: 10.1016/j.immuni.2022.07.014.Margeta MA, Yin Z, Madore C, et al. Apolipoprotein
E4 impairs the response of neurodegenerative retinal
microglia and prevents neuronal loss in glaucoma.
Immunity. 2022, 55(9): 1627-1644.e7. DOI: 10.1016/j.immuni.2022.07.014.
49、Wooff Y, Man SM, Aggio-Bruce R, et al. IL-1 family
members mediate cell death, inflammation and
angiogenesis in retinal degenerative diseases. Front
Immunol. 2019, 10: 1618. DOI: 10.3389/fimmu.2019.
01618.Wooff Y, Man SM, Aggio-Bruce R, et al. IL-1 family
members mediate cell death, inflammation and
angiogenesis in retinal degenerative diseases. Front
Immunol. 2019, 10: 1618. DOI: 10.3389/fimmu.2019.
01618.
50、Cavalli G, Colafrancesco S, Emmi G, et al. Interleukin
1α: a comprehensive review on the role of IL-1α in
the pathogenesis and treatment of autoimmune and
inflammatory diseases. Autoimmun Rev. 2021, 20(3):
102763. DOI: 10.1016/j.autrev.2021.102763.Cavalli G, Colafrancesco S, Emmi G, et al. Interleukin
1α: a comprehensive review on the role of IL-1α in
the pathogenesis and treatment of autoimmune and
inflammatory diseases. Autoimmun Rev. 2021, 20(3):
102763. DOI: 10.1016/j.autrev.2021.102763.
51、Chen M, Rong R, Xia X. Spotlight on pyroptosis: role in
pathogenesis and therapeutic potential of ocular diseases.
J Neuroinflammation., 2022, 19(1): 183. DOI: 10.1186/
s12974-022-02547-2.Chen M, Rong R, Xia X. Spotlight on pyroptosis: role in
pathogenesis and therapeutic potential of ocular diseases.
J Neuroinflammation., 2022, 19(1): 183. DOI: 10.1186/
s12974-022-02547-2.
52、Joshi A, Mayr M. In aptamers they trust: the caveats
of the SOMAscan biomarker discovery platform from
SomaLogic. Circulation. 2018, 138(22): 2482-2485. DOI:
10.1161/CIRCULATIONAHA.118.036823.Joshi A, Mayr M. In aptamers they trust: the caveats
of the SOMAscan biomarker discovery platform from
SomaLogic. Circulation. 2018, 138(22): 2482-2485. DOI:
10.1161/CIRCULATIONAHA.118.036823.